I ran into an issue recently using clusterR to predict randomForest
model outputs. It took me awhile to troubleshoot, so if you have had
the same problem, hopefully you find this post and it saves you some
time. If you’ve come up with a different solution (i.e. I’m wrong),
please let me know. It turns out I was wrong! See the solution below
In short: when using clusterR to predict the probability of occurrence output of a randomForest model, it automatically defaults to the binary output. This is problematic if you’re interested in the probability output to use in another RF model or if you want to set the threshold at which to convert the probability model to a binary data set. However, clusterR works just fine to predict other randomForest outputs (e.g. percent cover, type=’response’). See below for an example.
clusterR is a flexible interface for using cluster with other
functions. This function only works with functions that have a Raster*
object as first argument and that operate on a cell by cell basis (i.e.,
there is no effect of neigboring cells) and return an object with the
same number of cells as the input raster object (from ?clusterR). Martin
Šiklar has a nice
post
on increasing the speed of raster processing using clusterR.
Below I’ve generated a basic randomForest classification model and predict the probability of occurrence output (continuous).
library(raster)
//load an example raster stack
logo <- stack(system.file("external/rlogo.grd", package="raster"))
plot(logo)
//generate presence and random background (pseudo-absence) points
presence <- matrix(c(85, 60, 79, 50, 20,48, 50, 30, 70, 90, 48, 48, 48, 53, 50, 46, 54, 70, 84, 85, 74, 84, 95, 85, 66, 42, 26, 4, 19, 17, 5, 52, 10, 68, 50, 52, 18, 20, 30, 60, 7, 14, 26, 29, 39, 45, 51, 56, 46, 38, 31, 22, 34, 60, 70, 73, 63, 46, 43, 28), ncol=2)
//random background
background <- cbind(runif(250)*(xmax(logo)-xmin(logo))+xmin(logo), runif(250)*(ymax(logo)-ymin(logo))+ymin(logo))
//extract values for points from stack
xy <- rbind(cbind(1, presence), cbind(0, background))
v <- cbind(xy[,1], extract(logo, xy[,2:3]))
colnames(v)[1] <- 'presback'
//convert to dataframe
v.df<-as.data.frame(v)
//check the dataframe
typeof(v.df$presback) #double
as.factor(v.df$presback)
//random forests needs to see the '1' first thus the relevel; 1=present, 0=absent;
v.df$presback <- relevel(as.factor(ifelse(v.df$presback==1,1,0)),"1")
str(v.df) #ensure y (presback) is a factor because we're interested in a classification, not a regression
//run a random forest model
library(randomForest)
model.RF <- randomForest(presback~., data=v.df) #classification model
model.RF #note, it's not a great model fit, but not the point here
//predict probability output without cluster
r.prob <- predict(logo, type='prob', model.RF, progress='text') #type = 'prob' -> probability of occurrence model
plot(r.prob)
points(presence, bg='blue', pch=21)
points(background, bg='red', pch=21)
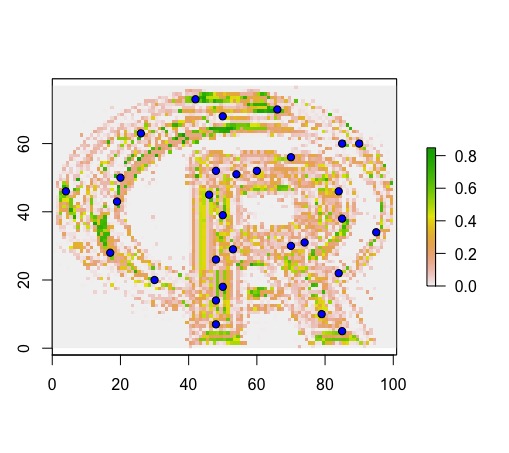
Next, I use clusterR to predict the same output; however, it results in a binary output.
//now make the prediction using multiple cores
library(cluster)
library(parallel)
//create a cluster and ID the # of cores to use
beginCluster(3)
//predict fxn using clusterR
r.prob.Cluster<-clusterR(logo, predict, args=list(model.RF), progress='text', type='prob')
endCluster() #delete the cluster
plot(r.prob.Cluster)
points(presence, bg='blue', pch=21)
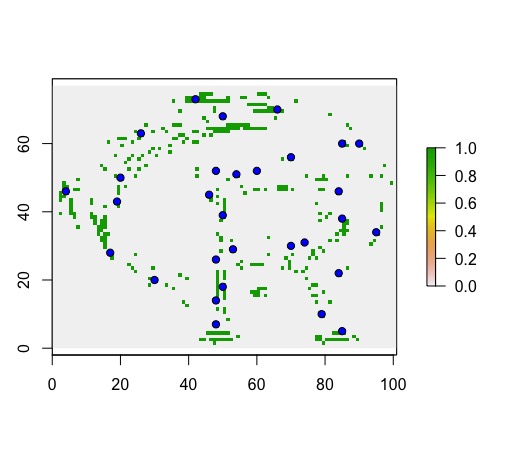
I’d highly recommend using clusterR whenever possible as it will save you a lot of time, particularly when making predictions with raster data. It will allow you to work in parallel and take advantage of multiple cores on your machine. My goal here is to simply make you aware of the issue with clusterR with respect to predicting probability of occurrence models. I am unaware of any solution, except to use the longhand predict function in the raster library when you need to predict the probability of occurrence model output.
Solution
I simply failed to have include type=’prob’ inside the args() part of the clusterR() function. Once I cleaned up the syntax, I had no problem!
Top image: Diamond Peak, Never Summer Range, Colorado, USA
